Research
Please visit the above links to have a more detailed look at our research projects. Below is a list of Centers that we are leading or affiliated with.
NSF MBM at UIUC - Miniature Brain Machinery
NSF STC EBICS at MIT/GT/UIUC - Emergent Behavior of Integrated Cellular Systems
NSF IGERT at UIUC - Cellular and Molecular Mechanics and Bionanotechnology
(download brochure)NIH Training Grant at UIUC - Midwestern Cancer Nanotechnology Training Center
(download brochure)NSF CiiT (I/UCRC) at UIUC - Center for Innovative Instrumentation Technology
NSF NSEC at OSU - Center for Affordable Nanoengineering for Polymeric Micro and Nanodevices
Research:
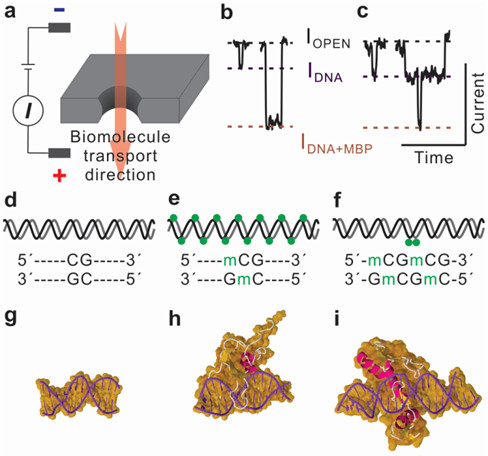
Detection and Quantification of DNA Methylation
DNA methylation, an epigenetic modification of a methyl group at the 5-carbon position of cytosine (5mC), has been found to be closely associated with carcinogenesis. Thus, genomic extracts holding this modification can be used as biomarker for sensitive detection of cancer, and these genomic DNAs can be collected from serum, plasma, urine, and stool. Nanopore-based direct methylation assay circumvents bisulfite conversion and PCR, suitable for screening of low sample volume obtained from genomic extracts. Methylated DNA is selectively labeled with methyl-binding proteins. Nanopore assay demonstrates discrimination of hypermethylated and unmethylated DNA on various length DNA fragments using sub 10 nm nanopores. Furthermore, low-methylated DNA fragments are differentiated from unmethylated DNA and the position of methylation is profiled using sensitive nanopore-based methylation assay.